Image Biomarkers
- Proprietary analysis for tumor texture and radiomics
- Development of imaging biomarkers for molecular profiling, prediction of response, prognosis, early detection of response and recurrence, hallmarks of cancer
- AI application and integration of imaging results with clinical outcome data for machine learning
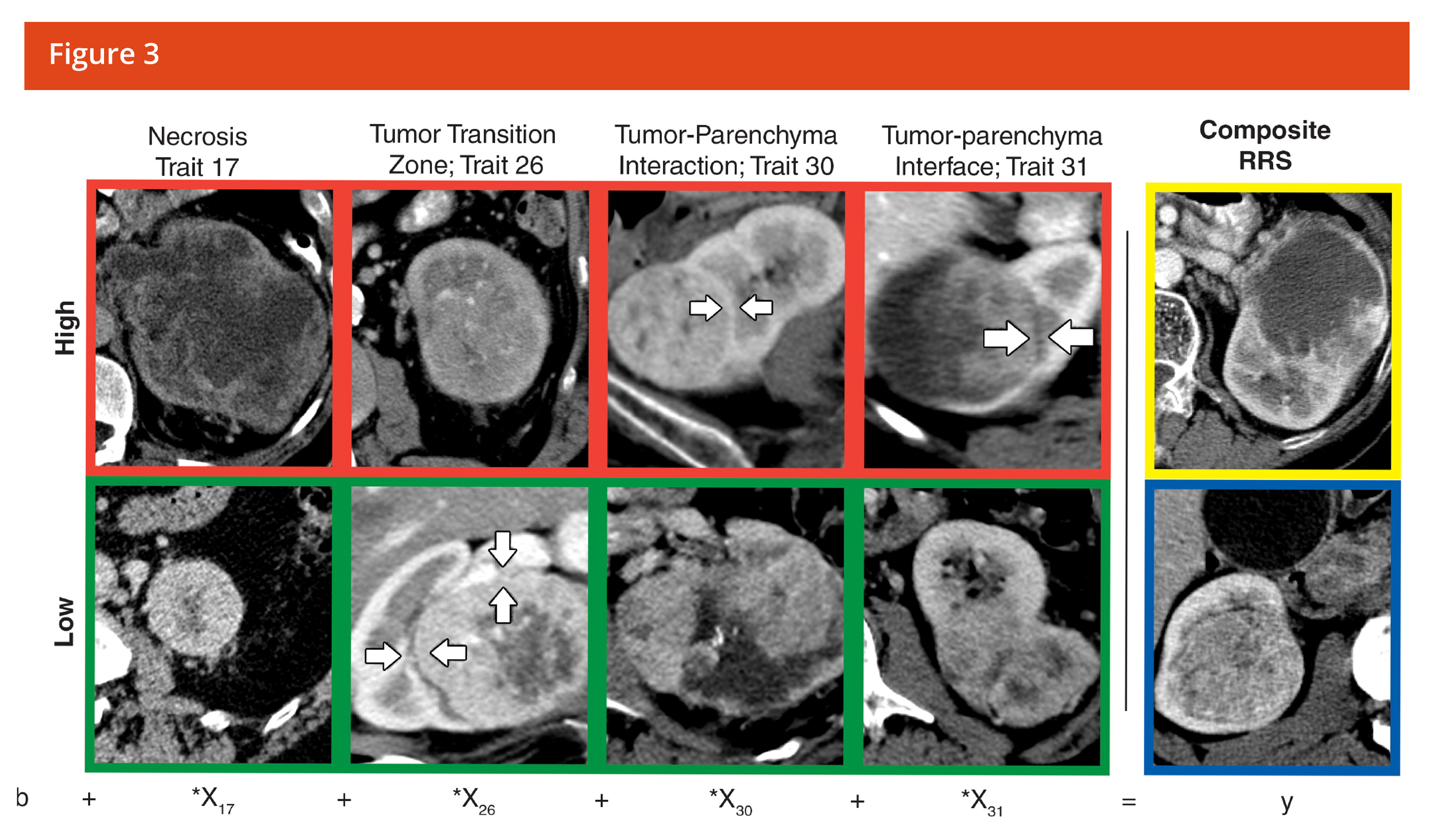
Quantitative Textural Analysis (QTA) is a post-processing technique that can be used to quantify changes in tissue by assessing the distribution of textural features within a tumor lesion and its change following treatment. QTA reduces the effect of photon noise and probes the biological diversity inherent in tumor complexity. QTA can quantify a range of measurable parameters based on enhancement characteristics and density changes on a pixel-by-pixel basis. QTA may be performed from standard of care CT, MRI, PET or Mammography scans. The system and platform required for measuring texture complexity is exclusively available at Imaging Endpoints’ Core Lab through a strategic partnership with TexRAD and is backed by over 50 publications in leading journals.
QTA measures density/intensity on a pixel-by-pixel basis. QTA creates a profile of pixel signal intensity over an area of interest on standard of care images
- Filter out photon noise
- Measure of signal intensity (SI) of each individual pixel
- Cluster pixels of similar SI together
- Mathematical transformation of histogram profile
- Representation of tumor morphology
- Characterization of tissue response and biology
QTA Quantitates and/or is predictive of the following:
- Heterogeneity
- Perfusion
- Hypoxia
- Genomics, transcriptomics, proteomics and metabolomics
- Other biological changes that may be indicative of response
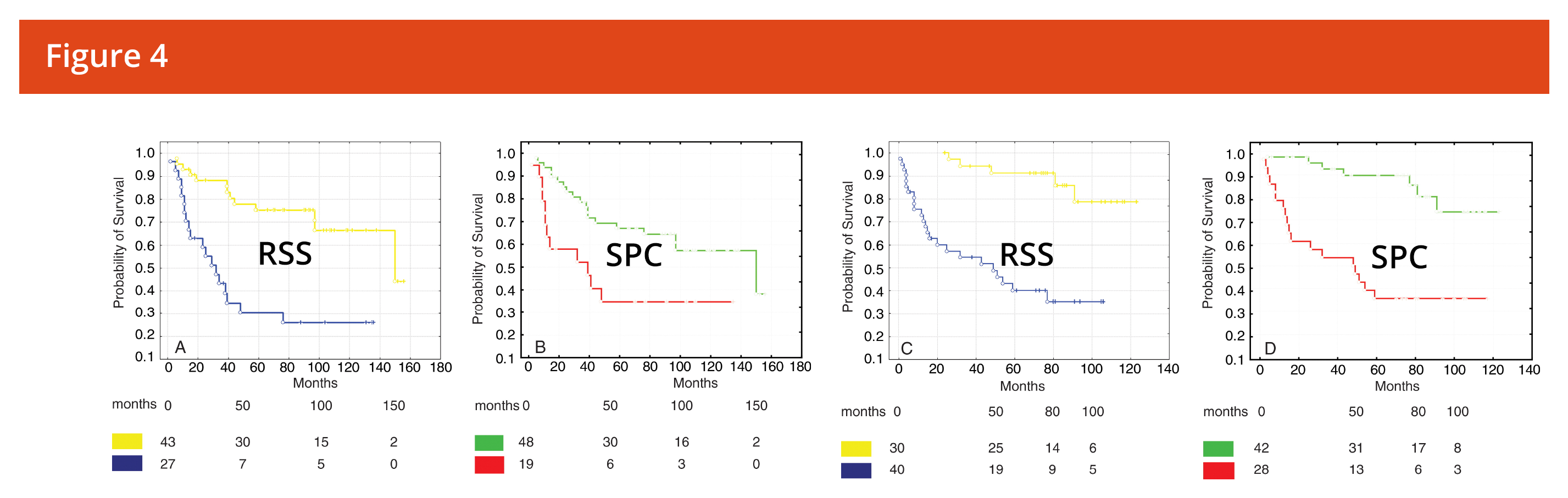
- Has been shown to be more predictive of response than RECIST
- Has been used to develop imaging biomarkers that predict response from baseline scans
Winner of the Alexander R Margulis Award for Scientific Excellence